Following the recent article arguing why PyPy is the future of Python, I must say, PyPy is not the future of Python, is the present. When I have tested it last time (PyPy-c 1.1.0) with Pyevolve into the optimization of a simple Sphere function, it was at least 2x slower than Unladen Swallow Q2, but in that time, PyPy was not able to JIT. Now, with this new release of PyPy and the JIT’ing support, the scenario has changed.
PyPy has evolved a lot (actually, you can see this evolution here), a nice work was done on the GC system, saving (when compared to CPython) 8 bytes per object allocated, which is very interesting for applications that makes heavy use of object allocation (GP system are a strong example of this, since when they are implemented on object oriented languages, each syntax tree node is an object). Efforts are also being done to improve support for CPython extensions (written in C/C++), one of them is a little tricky: the use of RPyC, to proxy through TCP the remote calls to CPython; but the other seems by far more effective, which is the creation of the CPyExt subsystem. By using CPyExt, all you need is to have your CPython API functions implemented in CPyExt, a lot of people is working on this right now and you can do it too, it’s a long road to have a good API coverage, but when you think about advantages, this road becomes small.
In order to benchmark CPython, Jython, CPython+Psyco, Unladen Swallow and PyPy, I’ve used the Rastrigin function optimization (an example of that implementation is here in the Example 7 of Pyevolve 0.6rc1):
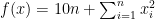
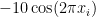
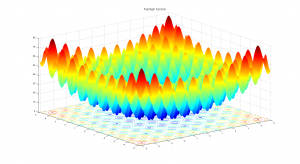
Due to its large search space and number of local minima, Rastrigin function is often used to measure the performance of Genetic Algorithms. Rastrigin function has a global minimum at  where the
where the 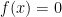 ; in order to increase the search space and required resources, I’ve used 40 variables (
; in order to increase the search space and required resources, I’ve used 40 variables ( ) and 10k generations.
) and 10k generations.
Here are the information about versions used in this benchmark:
- OS Ubuntu Linux 10.04 LTS (lucid)
- CPython 2.6.5 (Apr 16 2010)
- Unladen Swallow trunk (r1153)
- Jython 2.5.1 (Sun JVM 1.6.0_20, Server Mode)
- CPython 2.6.5 + PsycoV2 trunk (r74587)
- CPython 2.6.5 + Psyco 1.6.0 (default lucid package)
- PyPy trunk (r74537)
No warmup was performed in JVM or in PyPy. PyPy translator was executed using the “-Ojit” option in order to get the JIT version of the Python interpreter. The JVM was executed using the server mode, I’ve tested the client and server mode for Sun JVM and IcedTea6, the best results were observed from the server mode using Sun JVM, however when I’ve compared the client mode of IcedTea6 with the client mode of Sun JVM, the best results observed were from IcedTea6 (the same as using server mode in IcedTea6). Unladen Swallow was compiled using the project wiki instructions for building an optimized binary.
The machine used was an Intel(R) Core(TM) 2 Duo E4500 (2×2.20Ghz) with 2GB of RAM.
The result of the benchmark (measured using wall time) in seconds for each setup (these results are the best of 3 sequential runs):
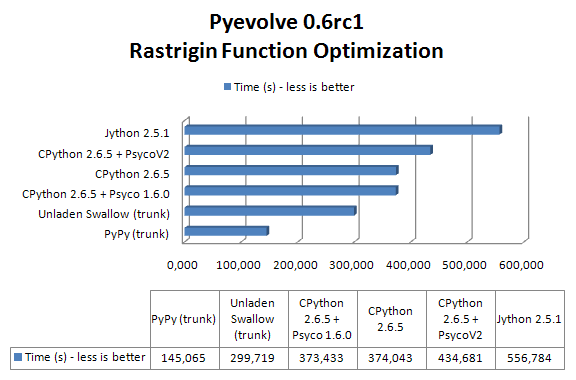
As you can see, PyPy with JIT got a speedup of 2.57x when compared to CPython 2.6.5 and 2.0x faster than Unladen Swallow current trunk.
PyPy is not only the future of Python, but is becoming the present right now. PyPy will not bring us only an implementation of Python in Python (which in itself is the valuable result of great efforts), but also will bring the performance back (which many doubted at the beginning, wondering how could it be possible for an implementation of Python in Python be faster than an implementation in C ? And here is where the translation and JIT magic enters). When the time comes that Python interpreter can be entire written in a high level language (actually almost the same language, which is really weird), Python community can put their focus on improving the language itself instead of spending time solving the complexity of the lower level languages, is this not the great point of those efforts ?
By the way, just to note, PyPy isn’t only a translator for the Python interpreter written in RPython, it’s a translator of RPython, what means that PyPy isn’t only the future of Python, but probably, the future of many interpreters.