I’m proud to announce that the new versions of Pyevolve will have Genetic Programming support; after some time fighting with these evil syntax trees, I think I have a very easy and flexible implementation of GP in Python. I was tired to see people giving up and trying to learn how to implement a simple GP using the hermetic libraries for C/C++ and Java (unfortunatelly I’m a Java web developer hehe).
The implementation is still under some tests and optimization, but it’s working nice, here is some details about it:
The implementation has been done in pure Python, so we still have many bonus from this, but unfortunatelly we lost some performance.
The GP core is very very flexible, because it compiles the GP Trees in Python bytecodes to speed the execution of the function. So, you can use even Python objects as terminals, or any possible Python expression. Any Python function can be used too, and you can use all power of Python to create those functions, which will be automatic detected by the framework using the name prefix =)
As you can see in the source-code, you don’t need to bind variables when calling the syntax tree of the individual, you simple use the “getCompiledCode” method which returns the Python compiled function ready to be executed.
Here is a source-code example:
from pyevolve import *
import math
error_accum = Util.ErrorAccumulator()
# This is the functions used by the GP core,
# Pyevolve will automatically detect them
# and the they number of arguments
def gp_add(a, b): return a+b
def gp_sub(a, b): return a-b
def gp_mul(a, b): return a*b
def gp_sqrt(a): return math.sqrt(abs(a))
def eval_func(chromosome):
global error_accum
error_accum.reset()
code_comp = chromosome.getCompiledCode()
for a in xrange(0, 5):
for b in xrange(0, 5):
# The eval will execute a pre-compiled syntax tree
# as a Python expression, and will automatically use
# the "a" and "b" variables (the terminals defined)
evaluated = eval(code_comp)
target = math.sqrt((a*a)+(b*b))
error_accum += (target, evaluated)
return error_accum.getRMSE()
def main_run():
genome = GTree.GTreeGP()
genome.setParams(max_depth=5, method="ramped")
genome.evaluator.set(eval_func)
ga = GSimpleGA.GSimpleGA(genome)
# This method will catch and use every function that
# begins with "gp", but you can also add them manually.
# The terminals are Python variables, you can use the
# ephemeral random consts too, using ephemeral:random.randint(0,2)
# for example.
ga.setParams(gp_terminals = ['a', 'b'],
gp_function_prefix = "gp")
# You can even use a function call as terminal, like "func()"
# and Pyevolve will use the result of the call as terminal
ga.setMinimax(Consts.minimaxType["minimize"])
ga.setGenerations(1000)
ga.setMutationRate(0.08)
ga.setCrossoverRate(1.0)
ga.setPopulationSize(2000)
ga.evolve(freq_stats=5)
print ga.bestIndividual()
if __name__ == "__main__":
main_run()
I’m very happy and testing the possibilities of this GP implementation in Python.
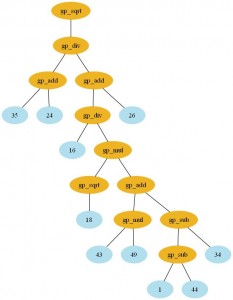
And of course, everything in Pyevolve can be visualized any time you want (click to enlarge):
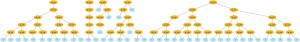

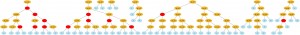
The visualization is very flexible too, if you use Python decorators to set how functions will be graphical represented, you can have many interesting visualization patterns. If I change the function “gp_add” to:
@GTree.gpdec(representation="+", color="red")
def gp_add(a, b): return a+b
We’ll got the follow visualization (click to enlarge):
I hope you enjoyed it, I’m currently fixing some bugs, implementing new features, docs and preparing the next release of Pyevolve, which will take some time yet =)