Update 17/01: reddit discussion thread.
Update 19/01: hacker news thread.
The codex
 The Voynich Manuscript is a hand-written codex written in an unknown system and carbon-dated to the early 15th century (1404–1438). Although the manuscript has been studied by some famous cryptographers of the World War I and II, nobody has deciphered it yet. The manuscript is known to be written in two different languages (Language A and Language B) and it is also known to be written by a group of people. The manuscript itself is always subject of a lot of different hypothesis, including the one that I like the most which is the “culture extinction” hypothesis, supported in 2014 by Stephen Bax. This hypothesis states that the codex isn’t ciphered, it states that the codex was just written in an unknown language that disappeared due to a culture extinction. In 2014, Stephen Bax proposed a provisional, partial decoding of the manuscript, the video of his presentation is very interesting and I really recommend you to watch if you like this codex. There is also a transcription of the manuscript done thanks to the hard-work of many folks working on it since many moons ago.
The Voynich Manuscript is a hand-written codex written in an unknown system and carbon-dated to the early 15th century (1404–1438). Although the manuscript has been studied by some famous cryptographers of the World War I and II, nobody has deciphered it yet. The manuscript is known to be written in two different languages (Language A and Language B) and it is also known to be written by a group of people. The manuscript itself is always subject of a lot of different hypothesis, including the one that I like the most which is the “culture extinction” hypothesis, supported in 2014 by Stephen Bax. This hypothesis states that the codex isn’t ciphered, it states that the codex was just written in an unknown language that disappeared due to a culture extinction. In 2014, Stephen Bax proposed a provisional, partial decoding of the manuscript, the video of his presentation is very interesting and I really recommend you to watch if you like this codex. There is also a transcription of the manuscript done thanks to the hard-work of many folks working on it since many moons ago.
Word vectors
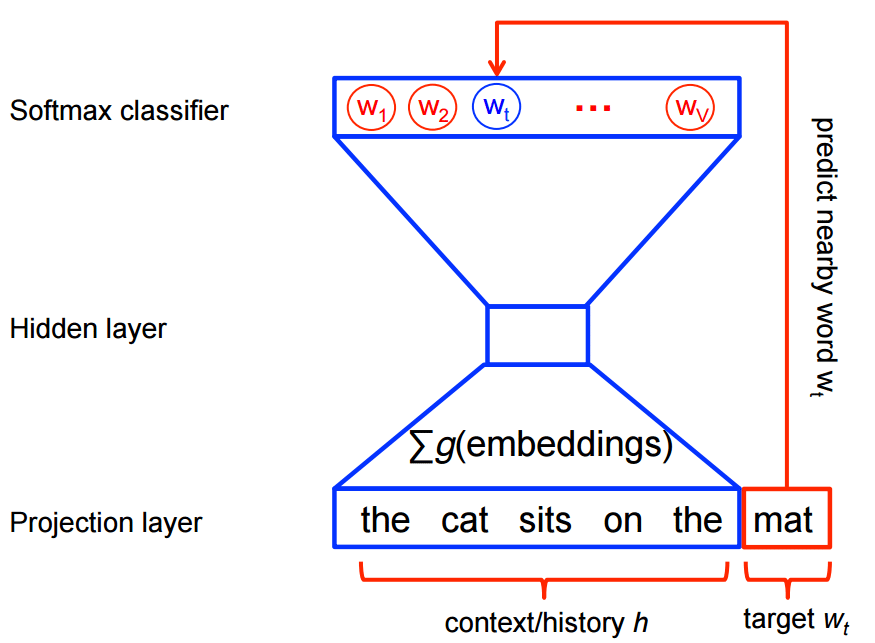
My idea when I heard about the work of Stephen Bax was to try to capture the patterns of the text using word2vec. Word embeddings are created by using a shallow neural network architecture. It is a unsupervised technique that uses supervided learning tasks to learn the linguistic context of the words. Here is a visualization of this architecture from the TensorFlow site:
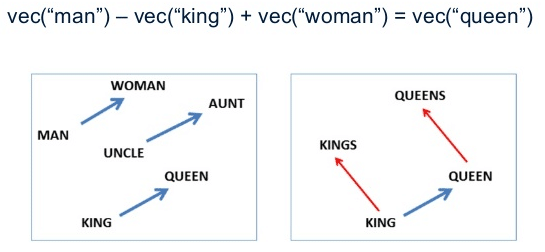
These word vectors, after trained, carry with them a lot of semantic meaning. For instance:
We can see that those vectors can be used in vector operations to extract information about the regularities of the captured linguistic semantics. These vectors also approximates same-meaning words together, allowing similarity queries like in the example below:
>>> model.most_similar("man")
[(u'woman', 0.6056041121482849), (u'guy', 0.4935004413127899), (u'boy', 0.48933547735214233), (u'men', 0.4632953703403473), (u'person', 0.45742249488830566), (u'lady', 0.4487500488758087), (u'himself', 0.4288588762283325), (u'girl', 0.4166809320449829), (u'his', 0.3853422999382019), (u'he', 0.38293731212615967)]
>>> model.most_similar("queen")
[(u'princess', 0.519856333732605), (u'latifah', 0.47644317150115967), (u'prince', 0.45914226770401), (u'king', 0.4466976821422577), (u'elizabeth', 0.4134873151779175), (u'antoinette', 0.41033703088760376), (u'marie', 0.4061327874660492), (u'stepmother', 0.4040161967277527), (u'belle', 0.38827288150787354), (u'lovely', 0.38668593764305115)]
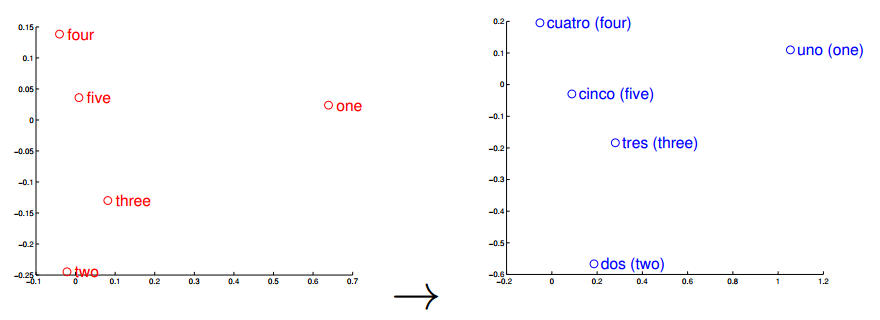
Word vectors can also be used (surprise) for translation, and this is the feature of the word vectors that I think that its most important when used to understand text where we know some of the words translations. I pretend to try to use the words found by Stephen Bax in the future to check if it is possible to capture some transformation that could lead to find similar structures with other languages. A nice visualization of this feature is the one below from the paper “Exploiting Similarities among Languages for Machine Translation“:
This visualization was made using gradient descent to optimize a linear transformation between the source and destination language word vectors. As you can see, the structure in Spanish is really close to the structure in English.
EVA Transcription
To train this model, I had to parse and extract the transcription from the EVA (European Voynich Alphabet) to be able to feed the Voynich sentences into the word2vec model. This EVA transcription has the following format:
<f1r.P1.1;H> fachys.ykal.ar.ataiin.shol.shory.cth!res.y.kor.sholdy!- <f1r.P1.1;C> fachys.ykal.ar.ataiin.shol.shory.cthorys.y.kor.sholdy!- <f1r.P1.1;F> fya!ys.ykal.ar.ytaiin.shol.shory.*k*!res.y!kor.sholdy!- <f1r.P1.1;N> fachys.ykal.ar.ataiin.shol.shory.cth!res.y,kor.sholdy!- <f1r.P1.1;U> fya!ys.ykal.ar.ytaiin.shol.shory.***!r*s.y.kor.sholdo*- # <f1r.P1.2;H> sory.ckhar.o!r.y.kair.chtaiin.shar.are.cthar.cthar.dan!- <f1r.P1.2;C> sory.ckhar.o.r.y.kain.shtaiin.shar.ar*.cthar.cthar.dan!- <f1r.P1.2;F> sory.ckhar.o!r!y.kair.chtaiin.shor.ar!.cthar.cthar.dana- <f1r.P1.2;N> sory.ckhar.o!r,y.kair.chtaiin.shar.are.cthar.cthar,dan!- <f1r.P1.2;U> sory.ckhar.o!r!y.kair.chtaiin.shor.ary.cthar.cthar.dan*-
The first data between “<” and “>” has information about the folio (page), line and author of the transcription. The transcription block above is the transcription for the first two lines of the first folio of the manuscript below:
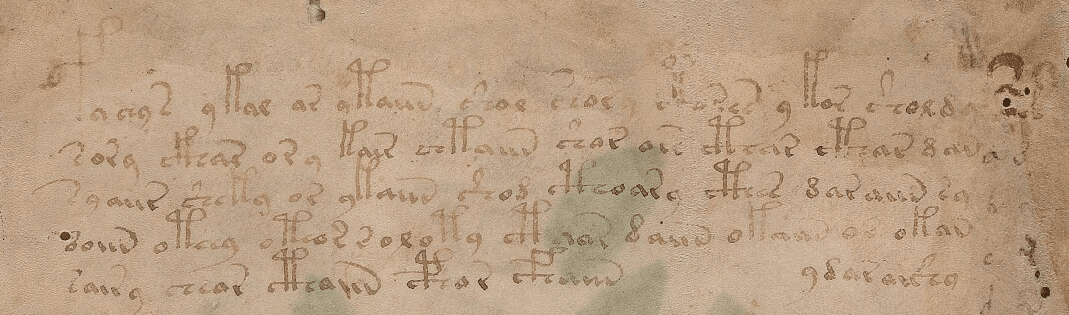
As you can see, the EVA contains some code characters, like for instance “!”, “*” and they all have some meaning, like to inform that the author doing that translation is not sure about the character in that position, etc. EVA also contains transcription from different authors for the same line of the folio.
To convert this transcription to sentences I used only lines where the authors were sure about the entire line and I used the first line where the line satisfied this condition. I also did some cleaning on the transcription to remove the drawings names from the text, like: “text.text.text-{plant}text” -> “text text texttext”.
After this conversion from the EVA transcript to sentences compatible with the word2vec model, I trained the model to provide 100-dimensional word vectors for the words of the manuscript.
Vector space visualizations using t-SNE
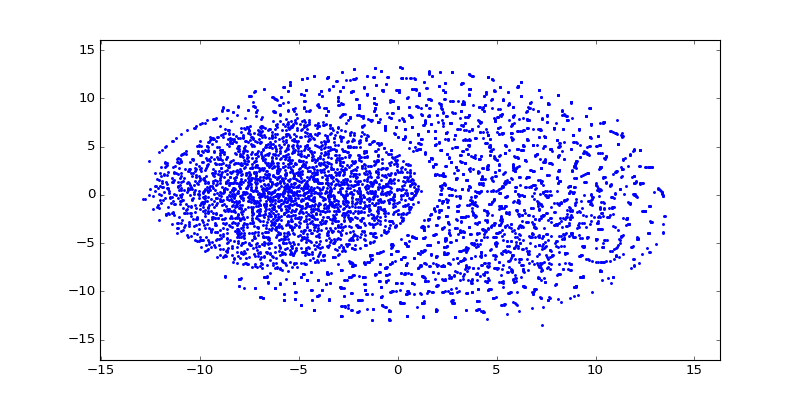
After training word vectors, I created a visualization of the 100-dimensional vectors into a 2D embedding space using t-SNE algorithm:
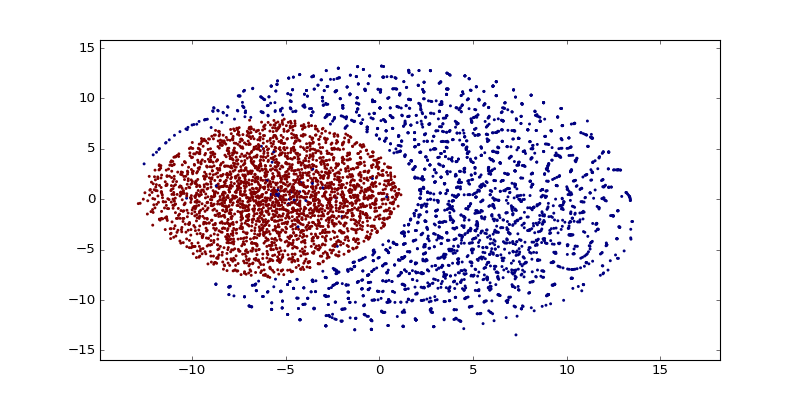
As you can see there are a lot of small clusters and there visually two big clusters, probably accounting for the two different languages used in the Codex (I still need to confirm this regarding the two languages aspect). After clustering it with DBSCAN (using the original word vectors, not the t-SNE transformed vectors), we can clearly see the two major clusters:
Now comes the really interesting and useful part of the word vectors, if use a star name from the folio below (it’s pretty obvious why it is know that this is probably a star name):
>>> w2v_model.most_similar("octhey")
[('qoekaiin', 0.6402825713157654),
('otcheody', 0.6389687061309814),
('ytchos', 0.566596269607544),
('ocphy', 0.5415685176849365),
('dolchedy', 0.5343093872070312),
('aiicthy', 0.5323750376701355),
('odchecthy', 0.5235849022865295),
('okeeos', 0.5187858939170837),
('cphocthy', 0.5159749388694763),
('oteor', 0.5050544738769531)]
I get really interesting similar words, like for instance the ocphy and other close star names:
It also returns the word “qoekaiin” from the folio 48, that precedes the same star name:
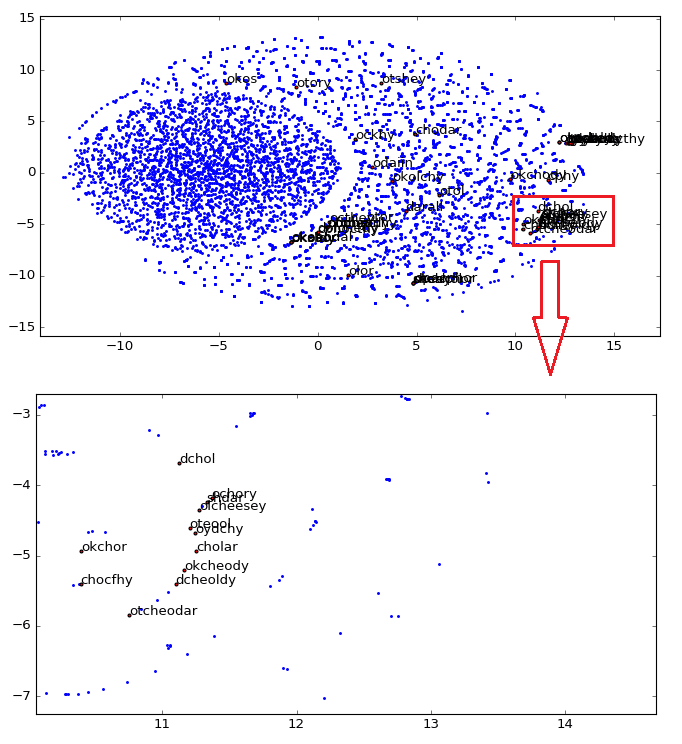
As you can see, word vectors are really useful to find some linguistic structures, we can also create another plot, showing how close are the star names in the 2D embedding space visualization created using t-SNE:
As you can see, we zoomed the major cluster of stars and we can see that they are really all grouped together in the vector space. These representations can be used for instance to infer plat names from the herbal section, etc.
My idea was to show how useful word vectors are to analyze unknown codex texts, I hope you liked and I hope that this could be somehow useful for other people how are also interested in this amazing manuscript.
– Christian S. Perone