Pyevolve benchmark on different Python flavors
So I did a comparative of Pyevolve GP/GA core in different Python interpreters. I’ve used my Pentium Core 2 Duo (E4500 @ 2.20GHz, 1GB RAM), using Ubuntu 9.04 and Windows XP SP3 just for IronPython 2.6.1 (IronPython doesn’t run with Mono, so I used the win xp with .net 2.0).
The interpreters used were:
I tried using 2009Q3 (the currently main trunk), but I think it’s unstable yet, cause it was more slow than 2009Q2, so I used 2009Q2; I compiled it with GCC 4.3.3 just using the default configure parameters (./configure).
I used the default CPython package of Ubuntu 9.04.
I used the default CPython package of Ubuntu 9.04 too, the python2.5 package.
I used the last svn version of the repository, the release 68612. My Pentium Core 2 Duo had only 1GB of RAM, and the PyPy translation process eats more RAM than Java (sorry for the joke), so I used a notebook with 3GB of RAM to create the pypy-c, what took 1 hour (I used –opt=3) and a beautiful ascii Mandelbrot fractal !
I used the default installer from the Jython project site. I used the Sun JRE 1.6.0_16.
I’ve used the 2.6 RC1 available at IronPython project site with MS .NET 2.0.
To test the GA core I’ve used this source-code (a simple sphere function):
from pyevolve import G1DList
from pyevolve import Mutators, Initializators
from pyevolve import GSimpleGA, Consts
# This is the Sphere Function
def sphere(xlist):
total = 0
for i in xlist:
total += i**2
return total
def run_main():
genome = G1DList.G1DList(140)
genome.setParams(rangemin=-5.12, rangemax=5.13)
genome.initializator.set(Initializators.G1DListInitializatorReal)
genome.mutator.set(Mutators.G1DListMutatorRealGaussian)
genome.evaluator.set(sphere)
ga = GSimpleGA.GSimpleGA(genome, seed=666)
ga.setMinimax(Consts.minimaxType["minimize"])
ga.setGenerations(1500)
ga.setMutationRate(0.01)
ga.evolve(freq_stats=500)
best = ga.bestIndividual()
if __name__ == "__main__":
run_main()
And to test the GP core, I’ve used this source-code (a simple symbolic regression):
from pyevolve import GTree
from pyevolve import Mutators
from pyevolve import GSimpleGA, Consts, Util
import math
rmse_accum = Util.ErrorAccumulator()
def gp_add(a, b): return a+b
def gp_sub(a, b): return a-b
def gp_mul(a, b): return a*b
def gp_sqrt(a): return math.sqrt(abs(a))
def eval_func(chromosome):
global rmse_accum
rmse_accum.reset()
code_comp = chromosome.getCompiledCode()
for a in xrange(0, 10):
for b in xrange(0, 10):
evaluated = eval(code_comp)
target = math.sqrt((a*a)+(b*b))
rmse_accum += (target, evaluated)
return rmse_accum.getRMSE()
def main_run():
genome = GTree.GTreeGP()
genome.setParams(max_depth=4, method="ramped")
genome.evaluator += eval_func
genome.mutator.set(Mutators.GTreeGPMutatorSubtree)
ga = GSimpleGA.GSimpleGA(genome, seed=666)
ga.setParams(gp_terminals = ['a', 'b'],
gp_function_prefix = "gp")
ga.setMinimax(Consts.minimaxType["minimize"])
ga.setGenerations(40)
ga.setCrossoverRate(1.0)
ga.setMutationRate(0.08)
ga.setPopulationSize(800)
ga(freq_stats=10)
best = ga.bestIndividual()
if __name__ == "__main__":
main_run()
UPDATE 19/08: the x-axis is measured in “seconds“, and the y-axis is the python flavor;
The results are are described in the graph below:
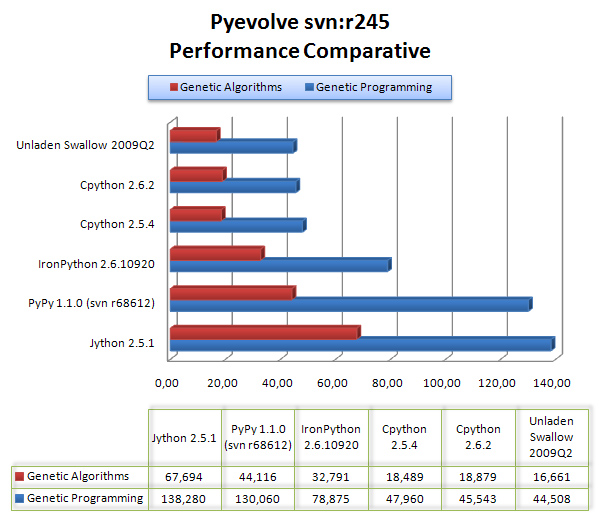 As we can see, Unladen Swallow 2009Q2 did a little better performance than CPython 2.6.2, but Jython and PyPy (experimental) were left behind in that scenario, even behind IronPython 2.6.1.
As we can see, Unladen Swallow 2009Q2 did a little better performance than CPython 2.6.2, but Jython and PyPy (experimental) were left behind in that scenario, even behind IronPython 2.6.1.

Der Christian,
you forgot to write down what your X axes (in this and the previous post) are. I assume it’s seconds, but it would be helpful if you explicitly wrote it down.
Oh it’s true hehe ! They are seconds as you noted, I’ll fix it ! Thanks Boris !
Do your measurements include startup time? Did you do a warmup phase?
Hello Philip ! They include startup time, the measure is the best of two runs.
So this benchmark ran for hours? If so, then I guess including startup time isn’t so important, and going through a proper warmup phase probably wouldn’t affect JVM/.NET times much either. Did you use Java’s server mode for Jython? (jython -J-server). It would default to slower client compiler for that kind of hardware
Philip, the x-axis is seconds, so it did not took hours to run. I’ll prepare a new benchmark using your comments, this time I’ll separate the startup time ! Thanks for comments.
Sorry, I’m misreading the commas in your results. Definitely omit startup time, and additionally you should do a warmup phase. For the JVM you want the warmup to aim for running your program’s inner loop something along the lines of ten thousand times. I’d recommend running the benchmark at least twice (maybe more, depending on pyevolve’s algorithms) as a warmup phase, then measuring the subsequent run in the same process. You want to ensure the JIT compiler has done its job.
You’re right, this is just a naive preliminary benchmark and doesn’t take the advantage of the possible performance (and this is specially true for jvm), but you must observe that this naive benchmark is more near of the real life cases, since nobody do a jvm warm up before running the ga/gp, they simple run. By the way, I’ll do another one benchmark with the warm up phase, maybe jvm can take advantage in a more hard ga/gp problem, since it will take more time and so the jit can do a better profile.
Very nice comparison! I’d definietly like to see results with startup times excluded and running for couple of minutes maybe so JIT compilers (.net have one also?) could do their work.
Thanks Naos, I’m planning a new comparison with a decent JVM warmup as asked by Philip Jenvey. The problem is that, in my point of view, it’s non-sense when we do this to solve a GA, why someone will solve an optimization problem (run it for some time to make jvm detect the best opt. for jitting the hotspot code) and then run it again to solve the problem ? I think that the time we spent warming up the jvm must be together with the total exection time, because in the real life, it this what happens.
>> because in the real life, it this what happens.
Depends on how you run your java program. Current main-stream JVM (pre-7) is not really command line application friendly. However, you can make all your java command-line application run faster by using tools like nailgun: http://martiansoftware.com/nailgun/background.html
So if I understood, you must leave a JVM running as background process ? The authors of nailgun seems a little confused: “What is it good for? I’m not sure yet.”.
I’m not testing the “most possible” performance of JVM or something like that, I’m testing performance of the framework in normal conditions. If we use nailgun to improve JVM performance, we can use Psyco for CPython too, so the benchmark becomes a “do all you can to improve performance”.
I agree, it wont make always sense to not include the warmup/startup time as some apps work only once so warmup becomes irrelevant. Also even if warmup does improve things considerably the user/programmer will have the above questions
1) how long before the warmup
2) how much speed increase after the warmup.
So a test with and without warmup is optimal.
The problem with these test is how variable the results are when the code changes. It makes it impossible to say for sure that there is an average in speed. But it is useful nonetheless to have a general idea how your programming language will respond in terms of speed , under specific conditions.
Personally I would want to see a test of how fast python performs when it calls code written in .NET, java or C/C++. A test I did with JAVA vs JYTHON shown me a 3-5x slowdown for jython. I would love to see my results verified or rejected. I think its the kind of speed test that python community misses.
Yep, sometimes the warmup/startup times are non-sense for some kind of applications like GAs/GPs, how can I imagine a man solving an optimization problem just to warmup the JVM, and then solve it again ?
The problem of using X method for speeding up, or X application to help in performance will create a combinatorial explosion for the benchmark, if I use X and Y for CPython for example, I must do a benchmark for: CPython alone, CPython with X, CPython with Y, CPython with X and Y, and so on… it’s not my goal to do a benchmark like that, I just want to show the behavior of different python flavors in a real life situation, because it’s the 90% of the cases.
Hey.
How about trying pypy 1.2 release which comes with JIT (or even svn trunk)? It should be much faster than 1.1 without JIT.
Hello Fijal =) This is my next post hehe, I’ve done some tests and I’m feeling that your implementation will be the faster among the other Python implementations. I’ll use the svn trunk, I just need to get time to prepare the benchmarks.
I thought I would try this in version 0.5 that I just installed, but there is no GTree in my egg (Pyevolve-0.5-py2.6.egg). Have you taken it out? I notice it is not documented.
The GTree entered in the 0.6 releases, you can use the 0.6rc1.
How did you run this test with pypy? I am using GTree, but it is not faster and it doesn’t work at all with Multi CPU, which means that it is slower when I use pypy. Is there a way to speed up the GTree performance?