Successful pyevolve multiprocessing speedup for Genetic Programming
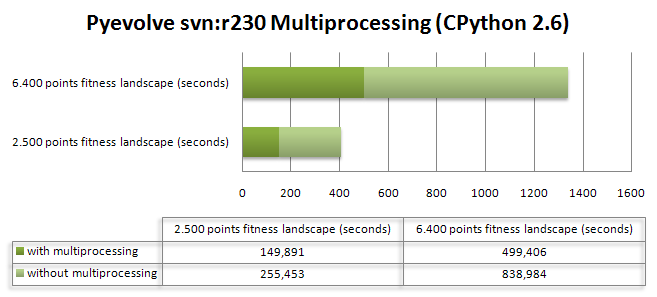
As we know, Genetic Programming usually requires intensive processing power for the fitness functions and tree manipulations (in crossover operations), and this fact can be a huge problem when using a pure Python approach like Pyevolve. So, to overcome this situation, I’ve used the Python multiprocessing features to implement a parallel fitness evaluation approach in Pyevolve and I was surprised by the super linear speedup I got for a cpu bound fitness function used to do the symbolic regression of the Pythagoras theorem: . I’ve used the same seed for the GP, so it has consumed nearly the same cpu resources for both test categories. Here are the results I obtained:
The first fitness landscape I’ve used had 2.500 points and the later had a fitness landscape of 6.400 points, here is the source code I’ve used (you just need to turn on the multiprocessing option using the setMultiProcessing method, so Pyevolve will use multiprocessing when you have more than one single core, you can enable the logging feature to check what’s going on behind the scenes):
from pyevolve import *
import math
rmse_accum = Util.ErrorAccumulator()
def gp_add(a, b): return a+b
def gp_sub(a, b): return a-b
def gp_mul(a, b): return a*b
def gp_sqrt(a): return math.sqrt(abs(a))
def eval_func(chromosome):
global rmse_accum
rmse_accum.reset()
code_comp = chromosome.getCompiledCode()
for a in xrange(0, 80):
for b in xrange(0, 80):
evaluated = eval(code_comp)
target = math.sqrt((a*a)+(b*b))
rmse_accum += (target, evaluated)
return rmse_accum.getRMSE()
def main_run():
genome = GTree.GTreeGP()
genome.setParams(max_depth=4, method="ramped")
genome.evaluator += eval_func
genome.mutator.set(Mutators.GTreeGPMutatorSubtree)
ga = GSimpleGA.GSimpleGA(genome, seed=666)
ga.setParams(gp_terminals = ['a', 'b'],
gp_function_prefix = "gp")
ga.setMinimax(Consts.minimaxType["minimize"])
ga.setGenerations(20)
ga.setCrossoverRate(1.0)
ga.setMutationRate(0.08)
ga.setPopulationSize(800)
ga.setMultiProcessing(True)
ga(freq_stats=5)
best = ga.bestIndividual()
if __name__ == "__main__":
main_run()
As you can see, the population size was 800 individuals with a 8% mutation rate and a 100% crossover rate for a simple 20 generations evolution. Of course you don’t need so many points in the fitness landscape, I’ve used 2.500+ points to create a cpu intensive fitness function, otherwise, the speedup can be less than 1.0 due the communication overhead between the processes. For the first case (2.500 points fitness landscape) I’ve got a 3.33x speedup and for the last case (6.400 points fitness landscape) I’ve got a 3.28x speedup. The tests were executed in a 2 cores pc (Intel Core 2 Duo).

Hello,
I wanted to give this multi-processing a try, but on my machine I can’t seem to locate the ga.setMultiProcessing() method?
GSimpleGA instance has no attribute ‘setMultiProcessing’
I believe these are the relevant parts of the code:
import csv
from pyevolve import *
from pyevolve import G2DList
from pyevolve import GSimpleGA
from pyevolve import Crossovers
from pyevolve import Mutators
from pyevolve import Selectors
from pyevolve import Statistics
from pyevolve import DBAdapters
from math import exp
from eval import eval
from numpy import *
import Gnuplot, Gnuplot.funcutils
genome = G2DList.G2DList(yearsToModel, len(sites) )
genome.setParams(rangemin=min(treatments), rangemax=max(treatments),gauss_mu=0, gauss_sigma=1)
genome.evaluator.set(eval_func)
genome.crossover.set(Crossovers.G2DListCrossoverSingleHPoint)
genome.mutator.set(Mutators.G2DListMutatorIntegerGaussian)
ga = GSimpleGA.GSimpleGA(genome, seed=333)
ga.selector.set(Selectors.GRouletteWheel)
ga.terminationCriteria.set(GSimpleGA.ConvergenceCriteria)
ga.setMultiProcessing(True)
Hello Alec, this feature is in the SVN trunk only, will be part of the 0.6 release, but you can just checkout the subversion repository of subversion. I’m expecting to release this new version in this month.
Multiprocessing is a good thing, but what about clusters? I hoped you would implement it using pp module.
Hello Senyai, in fact I’m implementing (it is working, I need to adapt some more new features and test again) an island model approach. But is in my plans to implement (for the 0.6 release) a coarse grained approach too. There is some people parallelizing Pyevolve using MPI too, but I don’t know the current state of this implementation. But is sure that the next step in Pyevolve roadmap, will be a stable distributed feature =)
Hi,
I would like to ask how progress is going on implementing this for clusters?
Hi,
I am trying to implement pyevolve to analyse experimental data using solutions of ODEs.
The evaluation of the fitness function is extremely costly and I need to parallelize as much as possible. It would be really nice to be able to distribute over a list of IPs.
I am also having problems setting ga.setMultiProcessing in mac OS Lion.
Thanks!
Sorry,
I just realize my comment can be missleading.
The code that it is published above runs perfectly well.
I am having problems setting multitasking features in Rosenbrock example.
Cheers,
Thanks for your useful work. But when I used pyevolve with multiprocessing, losts of python.exe are left in memory. I can’t kill them using the windows command ‘taskkill’. I need your help.
My ga setting as follows:
ga.setMultiProcessing(True)
ga.stepCallback.set(stepinfo)
My setCallback function is defined as follows:
def stepinfo(ga):
time.sleep(60)
pidid=os.getpid()
os.system(‘C:\\Windows\\System32\\taskkill.exe /FI “IMAGENAME eq python.exe” /FI “PID ne ‘
+str(pidid)+'” /F’)
time.sleep(60)