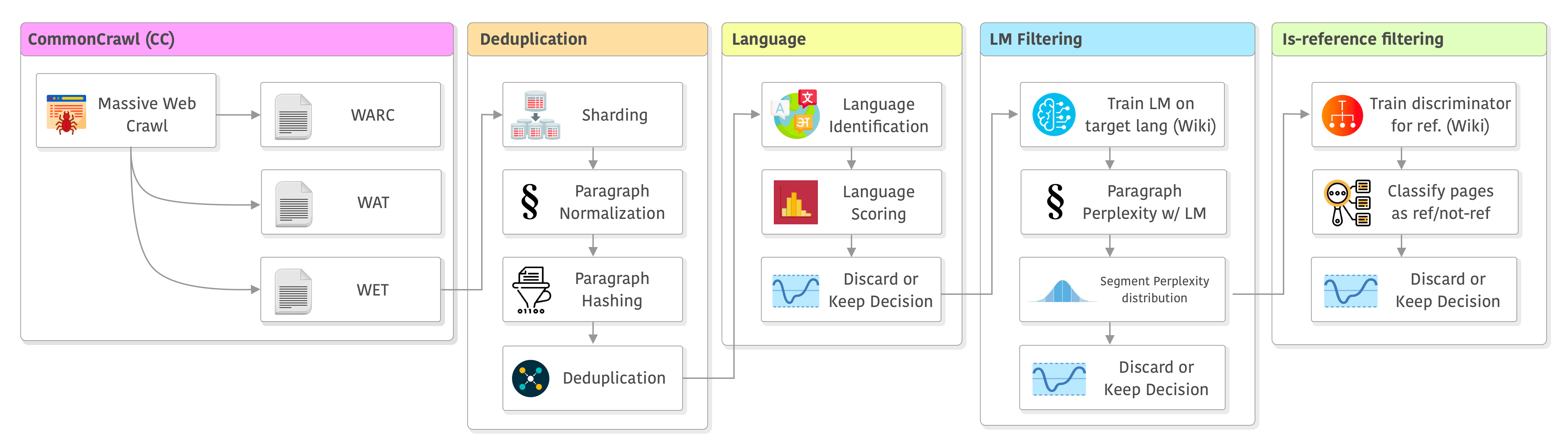
I really like to peek into different ML codebases for distributed training and this is a very short post on some things I found interesting in Torch Titan:
Disable and control of Python’s garbage collector (GC): titan codebase disables the Python GC and then manually forces a collection in the beginning of every training step during the training loop. This makes sense, but I’m not sure what are the gains of doing it, I think doing every step can be too much and I’m not sure if taking control of GC would be worth for the gains you get by manually controlling it, especially depending on complexity of other dependencies you use, as this could cause unintended behavior that would be difficult to associate with the GC collection;
Custom GPU memory monitoring: titan has a custom class to monitor GPU memory that is quite nice, it resets peak stats and empty the CUDA caching allocator upon initialization. At every step then they collect the peak stats for both small and large pools by capturing the stats for active, reserved and also failed retries and number of OOMs. It is very common for people to just monitor max GPU usage externally from NVML, however, this ignores the fact that PyTorch uses a caching allocator and that you need to look at the internal memory management mechanism inside PyTorch. If you don’t do that, you will certainly be mislead by what you are getting from NVML;
Custom profiling context manager: they wrote a context manager for profiling, where they measure time it takes to dump the profiling data per rank. Interesting here that there is a barrier at the end, which makes sense, but this is often the pain point of distributed training with PyTorch + NCCL;
Measuring data loading: this is of minor interest, but I liked the idea of not iterating on data loader in the loop statement itself but manually calling next() to get the batches, that makes it easier to measure data loading, that they average at the end for each epoch;
Logging MFU (model FLOPS utilization): they also compute and log MFU, which is quite helpful;
Delete predictions before backward: titan also deletes the model predictions before the backward() call to avoid memory peaks. This can be quite effective since you really don’t need this tensor anymore and you can delete it immediately before the backward pass;
Reduction of NCCL timeout: after the first training step, they reduce the NCCL timeout from the default 10 min to 100 sec. This is nice if you have well behaved replicas code and don’t need to do anything more complex, but 100 sec is a very short timeout that I would be careful using, it might be a good fit for your load but if your replicas drift a bit more, then you will need to keep adding barriers to avoid timeouts that can be incredibly difficult to debug and cause a lot of headaches;
Distributed checkpointing with mid-epoch checkpoint support: this is a very cool implementation, it uses distributed checkpointing from PyTorch. They create some wrappers (e.g. for optimizer) where they implement the Stateful protocol to support checkpointing. They also use the StatefulDataLoader from torchdata to do checkpointing of mid-epoch data loader state;
Misc: there are of course other interesting things, but it is cool to mention that they also implemented a no frills LLaMA model without relying on thousands of different libs (it seems it became fashionable nowadays to keep adding dependencies), so kudos for that to keep it simple.

 Just sharing ~100 slides about PyTorch 2 internals focusing on recent innovations (Dynamo, Inductor, and ExecuTorch). I had a lot of fun preparing this and hope you’ll enjoy it. I’m planning to record it soon.
Just sharing ~100 slides about PyTorch 2 internals focusing on recent innovations (Dynamo, Inductor, and ExecuTorch). I had a lot of fun preparing this and hope you’ll enjoy it. I’m planning to record it soon.


