Benford’s law is one of those very weird things that we can’t explain, and when we discover more and more phenomena that obey the law, we became astonished. Two people (Simon Newcomb – 1881 and Frank Benford – 1938) noted the law in the same way, while flipping pages of a logarithmic table book; they noticed that the pages at the beginning of the book were dirtier than the pages at the end.
Currently, there are no a priori criteria that say to us when a dataset will or will not obey the Benford’s Law. And it is because of this, that I’ve done an analysis on the Twitter Public Timeline.
The Twitter API to get Public Timeline is simply useless for this analysis because in the API Docs, they say that the Public Timeline is cached for 60 seconds ! 60 seconds is an eternity, and there is a request rating of 150 request/hour. So, it doesn’t help, buuuuuut, there is an alpha testing API with pretty and very useful streams of data from the Public Timeline; there are many methods in the Twitter Streaming API, and the most interesting one is the “Firehose”, which returns ALL the public statuses, but this method is only available for intere$ting people, and I’m not one of them. Buuuut, we have “Spritzer”, which returns a portion of all public statuses, since it’s only what we have available in the moment, it MUST be useful, and it’s a pretty stream of data =)
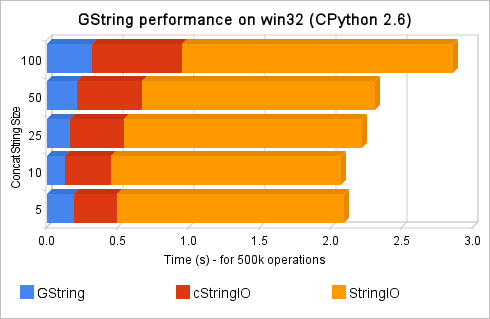
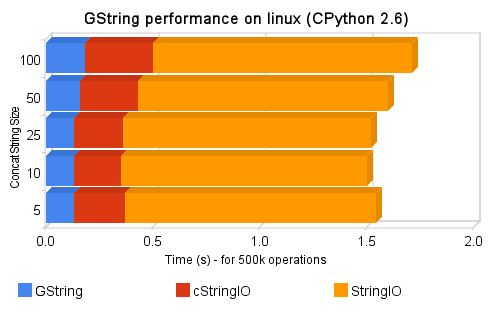
So, I’ve got the Spritzer real-time stream of data and processed each new status which arrived with a regex to find all the numbers in the status; if the status was “I have 5 dogs and 3 cats”, the numbers collected should be “[5, 3]”. All those accumulated numbers were then checked against the Benford’s Law. I’ve used Matplotlib to plot the two curves (the Benford’s Law and the Twitter statuses digits distribution) to empirically observe the correlation between them. You can note in the upper right corner of the video, the Pearson’s correlation between the two distributions too.
Here is the video (I’ve seen only after creating of the video, but the color of the curves are the inverse as seen in the legend):
The video represents the 15 minutes (3.160 captured statuses) of the Twitter Public Timeline. At the end of the video, you can see a Pearson’s correlation of 0.95. It seems that we have found another Benford’s son =)
The little tool to handle the Twitter Spritzer stream of data and plot the correlation graph in real-time was entirely written in Python, I’ll do a clean-up and post it here soon I got time. The tool has generated 1823 png images that were merged using ffmpeg.
I hope you enjoyed =)
UPDATE 11/08: the user “poobare” has cited an interesting paper about Benford’s Law on Reddit, here is the link.
More posts about Benford’s Law
Prime Numbers and the Benford’s Law